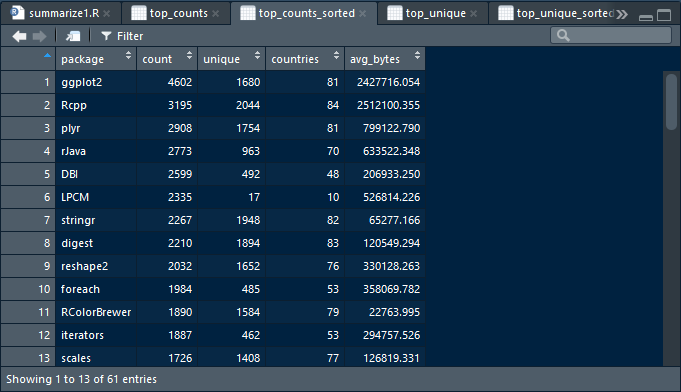
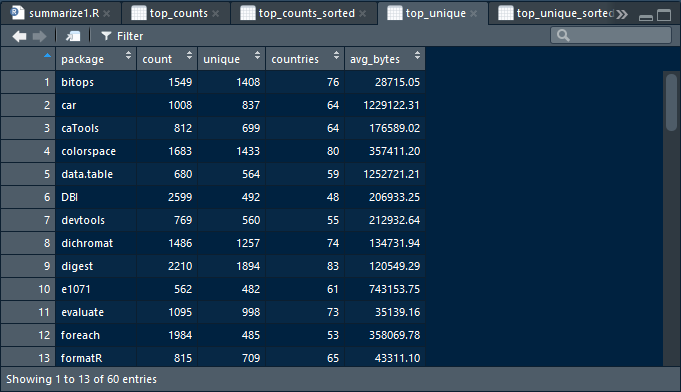
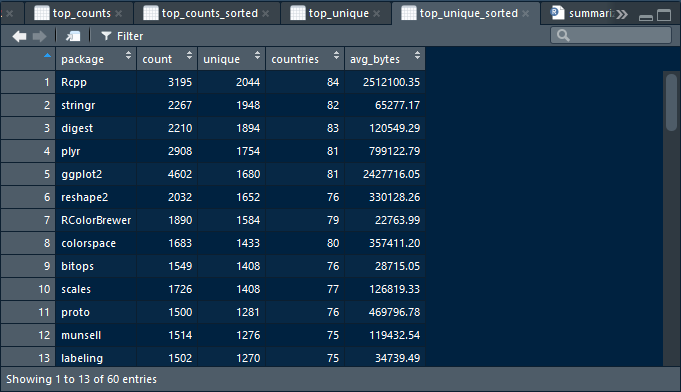
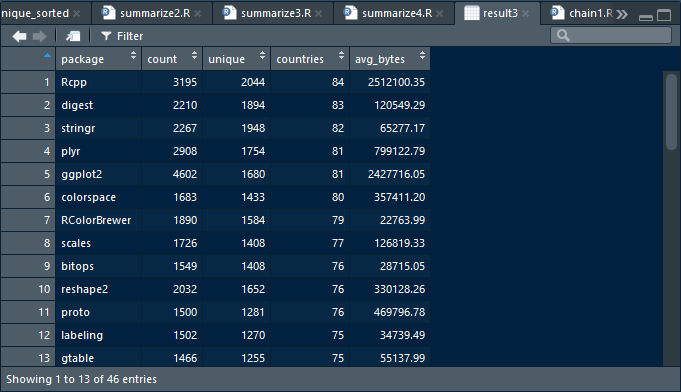
swirl()
| Welcome to swirl! Please sign in. If you've been here before, use the same name as you
| did then. If you are new, call yourself something unique.
What shall I call you? Krishnakanth Allika
| Please choose a course, or type 0 to exit swirl.
1: Getting and Cleaning Data
2: R Programming
3: Take me to the swirl course repository!
Selection: 1
| Please choose a lesson, or type 0 to return to course menu.
1: Manipulating Data with dplyr
2: Grouping and Chaining with dplyr
3: Tidying Data with tidyr
4: Dates and Times with lubridate
Selection: 3
| Attempting to load lesson dependencies...
| This lesson requires the ‘readr’ package. Would you like me to install it for you now?
1: Yes
2: No
Selection: 1
| Trying to install package ‘readr’ now...
also installing the dependencies ‘hms’, ‘clipr’
package ‘hms’ successfully unpacked and MD5 sums checked
package ‘clipr’ successfully unpacked and MD5 sums checked
package ‘readr’ successfully unpacked and MD5 sums checked
| Package ‘readr’ loaded correctly!
| This lesson requires the ‘tidyr’ package. Would you like me to install it for you now?
1: Yes
2: No
Selection: 1
| Trying to install package ‘tidyr’ now...
package ‘tidyr’ successfully unpacked and MD5 sums checked
| Package ‘tidyr’ loaded correctly!
| Package ‘dplyr’ loaded correctly!
| | 0%
| In this lesson, you'll learn how to tidy your data with the tidyr package.
...
|= | 2%
| Parts of this lesson will require the use of dplyr. If you don't have a basic knowledge
| of dplyr, you should exit this lesson and begin with the dplyr lessons from earlier in
| the course.
...
|=== | 4%
| tidyr was automatically installed (if necessary) and loaded when you started this
| lesson. Just to build the habit, (re)load the package with library(tidyr).
library(tidyr)
| That's a job well done!
|==== | 5%
| The author of tidyr, Hadley Wickham, discusses his philosophy of tidy data in his 'Tidy
| Data' paper:
|
| http://vita.had.co.nz/papers/tidy-data.pdf
|
| This paper should be required reading for anyone who works with data, but it's not
| required in order to complete this lesson.
...
|====== | 7%
| Tidy data is formatted in a standard way that facilitates exploration and analysis and
| works seamlessly with other tidy data tools. Specifically, tidy data satisfies three
| conditions:
|
| 1) Each variable forms a column
|
| 2) Each observation forms a row
|
| 3) Each type of observational unit forms a table
...
|======= | 9%
| Any dataset that doesn't satisfy these conditions is considered 'messy' data.
| Therefore, all of the following are characteristics of messy data, EXCEPT...
1: Variables are stored in both rows and columns
2: Column headers are values, not variable names
3: Every column contains a different variable
4: Multiple types of observational units are stored in the same table
5: Multiple variables are stored in one column
6: A single observational unit is stored in multiple tables
Selection: 3
| Keep up the great work!
|========= | 11%
| The incorrect answers to the previous question are the most common symptoms of messy
| data. Let's work through a simple example of each of these five cases, then tidy some
| real data.
...
|========== | 13%
| The first problem is when you have column headers that are values, not variable names.
| I've created a simple dataset called 'students' that demonstrates this scenario. Type
| students to take a look.
play()
| Entering play mode. Experiment as you please, then type nxt() when you are ready to
| resume the lesson.
download.file("http://vita.had.co.nz/papers/tidy-data.pdf","tidy-data.pdf")
trying URL 'http://vita.had.co.nz/papers/tidy-data.pdf'
Content type 'application/pdf' length 360450 bytes (352 KB)
downloaded 352 KB
nxt()
| Resuming lesson...
| The first problem is when you have column headers that are values, not variable names.
| I've created a simple dataset called 'students' that demonstrates this scenario. Type
| students to take a look.
students
grade male female
1 A 5 3
2 B 4 1
3 C 8 6
4 D 4 5
5 E 5 5
| That's a job well done!
|============ | 15%
| The first column represents each of five possible grades that students could receive
| for a particular class. The second and third columns give the number of male and female
| students, respectively, that received each grade.
...
|============= | 16%
| This dataset actually has three variables: grade, sex, and count. The first variable,
| grade, is already a column, so that should remain as it is. The second variable, sex,
| is captured by the second and third column headings. The third variable, count, is the
| number of students for each combination of grade and sex.
...
|=============== | 18%
| To tidy the students data, we need to have one column for each of these three
| variables. We'll use the gather() function from tidyr to accomplish this. Pull up the
| documentation for this function with ?gather.
?gather
| Nice work!
|================ | 20%
| Using the help file as a guide, call gather() with the following arguments (in order):
| students, sex, count, -grade. Note the minus sign before grade, which says we want to
| gather all columns EXCEPT grade.
gather(students,sex,count,-grade)
grade sex count
1 A male 5
2 B male 4
3 C male 8
4 D male 4
5 E male 5
6 A female 3
7 B female 1
8 C female 6
9 D female 5
10 E female 5
| All that hard work is paying off!
|================= | 22%
| Each row of the data now represents exactly one observation, characterized by a unique
| combination of the grade and sex variables. Each of our variables (grade, sex, and
| count) occupies exactly one column. That's tidy data!
...
|=================== | 24%
| It's important to understand what each argument to gather() means. The data argument,
| students, gives the name of the original dataset. The key and value arguments -- sex
| and count, respectively -- give the column names for our tidy dataset. The final
| argument, -grade, says that we want to gather all columns EXCEPT the grade column
| (since grade is already a proper column variable.)
...
|==================== | 25%
| The second messy data case we'll look at is when multiple variables are stored in one
| column. Type students2 to see an example of this.
students2
grade male_1 female_1 male_2 female_2
1 A 7 0 5 8
2 B 4 0 5 8
3 C 7 4 5 6
4 D 8 2 8 1
5 E 8 4 1 0
| Excellent work!
|====================== | 27%
| This dataset is similar to the first, except now there are two separate classes, 1 and
| 2, and we have total counts for each sex within each class. students2 suffers from the
| same messy data problem of having column headers that are values (male_1, female_1,
| etc.) and not variable names (sex, class, and count).
...
|======================= | 29%
| However, it also has multiple variables stored in each column (sex and class), which is
| another common symptom of messy data. Tidying this dataset will be a two step process.
...
|========================= | 31%
| Let's start by using gather() to stack the columns of students2, like we just did with
| students. This time, name the 'key' column sex_class and the 'value' column count. Save
| the result to a new variable called res. Consult ?gather again if you need help.
res<-gather(students2,sex_class,count,-grade)
| Your dedication is inspiring!
|========================== | 33%
| Print res to the console to see what we accomplished.
res
grade sex_class count
1 A male_1 7
2 B male_1 4
3 C male_1 7
4 D male_1 8
5 E male_1 8
6 A female_1 0
7 B female_1 0
8 C female_1 4
9 D female_1 2
10 E female_1 4
11 A male_2 5
12 B male_2 5
13 C male_2 5
14 D male_2 8
15 E male_2 1
16 A female_2 8
17 B female_2 8
18 C female_2 6
19 D female_2 1
20 E female_2 0
| Your dedication is inspiring!
|============================ | 35%
| That got us half way to tidy data, but we still have two different variables, sex and
| class, stored together in the sex_class column. tidyr offers a convenient separate()
| function for the purpose of separating one column into multiple columns. Pull up the
| help file for separate() now.
?separate
| You got it right!
|============================= | 36%
| Call separate() on res to split the sex_class column into sex and class. You only need
| to specify the first three arguments: data = res, col = sex_class, into = c("sex",
| "class"). You don't have to provide the argument names as long as they are in the
| correct order.
separate(data=res,col=sex_class,into=c("sex","class"))
grade sex class count
1 A male 1 7
2 B male 1 4
3 C male 1 7
4 D male 1 8
5 E male 1 8
6 A female 1 0
7 B female 1 0
8 C female 1 4
9 D female 1 2
10 E female 1 4
11 A male 2 5
12 B male 2 5
13 C male 2 5
14 D male 2 8
15 E male 2 1
16 A female 2 8
17 B female 2 8
18 C female 2 6
19 D female 2 1
20 E female 2 0
| You are amazing!
|=============================== | 38%
| Conveniently, separate() was able to figure out on its own how to separate the
| sex_class column. Unless you request otherwise with the 'sep' argument, it splits on
| non-alphanumeric values. In other words, it assumes that the values are separated by
| something other than a letter or number (in this case, an underscore.)
...
|================================ | 40%
| Tidying students2 required both gather() and separate(), causing us to save an
| intermediate result (res). However, just like with dplyr, you can use the %>% operator
| to chain multiple function calls together.
...
|================================= | 42%
| I've opened an R script for you to give this a try. Follow the directions in the
| script, then save the script and type submit() at the prompt when you are ready. If you
| get stuck and want to start over, you can type reset() to reset the script to its
| original state.
{r}
# Repeat your calls to gather() and separate(), but this time
# use the %>% operator to chain the commands together without
# storing an intermediate result.
#
# If this is your first time seeing the %>% operator, check
# out ?chain, which will bring up the relevant documentation.
# You can also look at the Examples section at the bottom
# of ?gather and ?separate.
#
# The main idea is that the result to the left of %>%
# takes the place of the first argument of the function to
# the right. Therefore, you OMIT THE FIRST ARGUMENT to each
# function.
#
students2 %>%
gather(sex_class ,count ,-grade ) %>%
separate(sex_class , c("sex", "class")) %>%
print
submit()
| Sourcing your script...
grade sex class count
1 A male 1 7
2 B male 1 4
3 C male 1 7
4 D male 1 8
5 E male 1 8
6 A female 1 0
7 B female 1 0
8 C female 1 4
9 D female 1 2
10 E female 1 4
11 A male 2 5
12 B male 2 5
13 C male 2 5
14 D male 2 8
15 E male 2 1
16 A female 2 8
17 B female 2 8
18 C female 2 6
19 D female 2 1
20 E female 2 0
| Excellent work!
|=================================== | 44%
| A third symptom of messy data is when variables are stored in both rows and columns.
| students3 provides an example of this. Print students3 to the console.
students3
name test class1 class2 class3 class4 class5
1 Sally midterm A B
2 Sally final C C
3 Jeff midterm D A
4 Jeff final E C
5 Roger midterm C B
6 Roger final A A
7 Karen midterm C A
8 Karen final C A
9 Brian midterm B A
10 Brian final B C
| You got it right!
|==================================== | 45%
| In students3, we have midterm and final exam grades for five students, each of whom
| were enrolled in exactly two of five possible classes.
...
|====================================== | 47%
| The first variable, name, is already a column and should remain as it is. The headers
| of the last five columns, class1 through class5, are all different values of what
| should be a class variable. The values in the test column, midterm and final, should
| each be its own variable containing the respective grades for each student.
...
|======================================= | 49%
| This will require multiple steps, which we will build up gradually using %>%. Edit the
| R script, save it, then type submit() when you are ready. Type reset() to reset the
| script to its original state.
{r}
# Call gather() to gather the columns class1
# through class5 into a new variable called class.
# The 'key' should be class, and the 'value'
# should be grade.
#
# tidyr makes it easy to reference multiple adjacent
# columns with class1:class5, just like with sequences
# of numbers.
#
# Since each student is only enrolled in two of
# the five possible classes, there are lots of missing
# values (i.e. NAs). Use the argument na.rm = TRUE
# to omit these values from the final result.
#
# Remember that when you're using the %>% operator,
# the value to the left of it gets inserted as the
# first argument to the function on the right.
#
# Consult ?gather and/or ?chain if you get stuck.
#
students3 %>%
gather(class ,grade , class1:class5 ,na.rm = TRUE) %>%
print
submit()
| Sourcing your script...
name test class grade
1 Sally midterm class1 A
2 Sally final class1 C
9 Brian midterm class1 B
10 Brian final class1 B
13 Jeff midterm class2 D
14 Jeff final class2 E
15 Roger midterm class2 C
16 Roger final class2 A
21 Sally midterm class3 B
22 Sally final class3 C
27 Karen midterm class3 C
28 Karen final class3 C
33 Jeff midterm class4 A
34 Jeff final class4 C
37 Karen midterm class4 A
38 Karen final class4 A
45 Roger midterm class5 B
46 Roger final class5 A
49 Brian midterm class5 A
50 Brian final class5 C
| That's correct!
|========================================= | 51%
| The next step will require the use of spread(). Pull up the documentation for spread()
| now.
?spread
| Keep working like that and you'll get there!
|========================================== | 53%
| Edit the R script, then save it and type submit() when you are ready. Type reset() to
| reset the script to its original state.
{r}
# This script builds on the previous one by appending
# a call to spread(), which will allow us to turn the
# values of the test column, midterm and final, into
# column headers (i.e. variables).
#
# You only need to specify two arguments to spread().
# Can you figure out what they are? (Hint: You don't
# have to specify the data argument since we're using
# the %>% operator.
#
students3 %>%
gather(class, grade, class1:class5, na.rm = TRUE) %>%
spread( test, grade) %>%
print
submit()
| Sourcing your script...
name class final midterm
1 Brian class1 B B
2 Brian class5 C A
3 Jeff class2 E D
4 Jeff class4 C A
5 Karen class3 C C
6 Karen class4 A A
7 Roger class2 A C
8 Roger class5 A B
9 Sally class1 C A
10 Sally class3 C B
| All that practice is paying off!
|============================================ | 55%
| readr is required for certain data manipulations, such as `parse_number(), which will
| be used in the next question. Let's, (re)load the package with library(readr).
library(readr)
| Nice work!
|============================================= | 56%
| Lastly, we want the values in the class column to simply be 1, 2, ..., 5 and not
| class1, class2, ..., class5. We can use the parse_number() function from readr to
| accomplish this. To see how it works, try parse_number("class5").
parse_number("class5")
[1] 5
| Your dedication is inspiring!
|=============================================== | 58%
| Now, the final step. Edit the R script, then save it and type submit() when you are
| ready. Type reset() to reset the script to its original state.
{r}
# We want the values in the class columns to be
# 1, 2, ..., 5 and not class1, class2, ..., class5.
#
# Use the mutate() function from dplyr along with
# parse_number(). Hint: You can "overwrite" a column
# with mutate() by assigning a new value to the existing
# column instead of creating a new column.
#
# Check out ?mutate and/or ?parse_number if you need
# a refresher.
#
students3 %>%
gather(class, grade, class1:class5, na.rm = TRUE) %>%
spread(test, grade) %>%
### Call to mutate() goes here %>%
mutate(class=parse_number(class)) %>%
print
submit()
| Sourcing your script...
name class final midterm
1 Brian 1 B B
2 Brian 5 C A
3 Jeff 2 E D
4 Jeff 4 C A
5 Karen 3 C C
6 Karen 4 A A
7 Roger 2 A C
8 Roger 5 A B
9 Sally 1 C A
10 Sally 3 C B
| You got it!
|================================================ | 60%
| The fourth messy data problem we'll look at occurs when multiple observational units
| are stored in the same table. students4 presents an example of this. Take a look at the
| data now.
students4
id name sex class midterm final
1 168 Brian F 1 B B
2 168 Brian F 5 A C
3 588 Sally M 1 A C
4 588 Sally M 3 B C
5 710 Jeff M 2 D E
6 710 Jeff M 4 A C
7 731 Roger F 2 C A
8 731 Roger F 5 B A
9 908 Karen M 3 C C
10 908 Karen M 4 A A
| You are doing so well!
|================================================= | 62%
| students4 is almost the same as our tidy version of students3. The only difference is
| that students4 provides a unique id for each student, as well as his or her sex (M =
| male; F = female).
...
|=================================================== | 64%
| At first glance, there doesn't seem to be much of a problem with students4. All columns
| are variables and all rows are observations. However, notice that each id, name, and
| sex is repeated twice, which seems quite redundant. This is a hint that our data
| contains multiple observational units in a single table.
...
|==================================================== | 65%
| Our solution will be to break students4 into two separate tables -- one containing
| basic student information (id, name, and sex) and the other containing grades (id,
| class, midterm, final).
|
| Edit the R script, save it, then type submit() when you are ready. Type reset() to
| reset the script to its original state.
{r}
# Complete the chained command below so that we are
# selecting the id, name, and sex column from students4
# and storing the result in student_info.
#
student_info <- students4 %>%
select(id ,name ,sex ) %>%
print
submit()
| Sourcing your script...
id name sex
1 168 Brian F
2 168 Brian F
3 588 Sally M
4 588 Sally M
5 710 Jeff M
6 710 Jeff M
7 731 Roger F
8 731 Roger F
9 908 Karen M
10 908 Karen M
| You are amazing!
|====================================================== | 67%
| Notice anything strange about student_info? It contains five duplicate rows! See the
| script for directions on how to fix this. Save the script and type submit() when you
| are ready, or type reset() to reset the script to its original state.
{r}
# Add a call to unique() below, which will remove
# duplicate rows from student_info.
#
# Like with the call to the print() function below,
# you can omit the parentheses after the function name.
# This is a nice feature of %>% that applies when
# there are no additional arguments to specify.
#
student_info <- students4 %>%
select(id, name, sex) %>%
### Your code here %>%
unique() %>%
print
submit()
| Sourcing your script...
id name sex
1 168 Brian F
3 588 Sally M
5 710 Jeff M
7 731 Roger F
9 908 Karen M
| Excellent job!
|======================================================= | 69%
| Now, using the script I just opened for you, create a second table called gradebook
| using the id, class, midterm, and final columns (in that order).
|
| Edit the R script, save it, then type submit() when you are ready. Type reset() to
| reset the script to its original state.
{r}
# select() the id, class, midterm, and final columns
# (in that order) and store the result in gradebook.
#
gradebook <- students4 %>%
### Your code here %>%
select(id,class,midterm,final) %>%
print
submit()
| Sourcing your script...
id class midterm final
1 168 1 B B
2 168 5 A C
3 588 1 A C
4 588 3 B C
5 710 2 D E
6 710 4 A C
7 731 2 C A
8 731 5 B A
9 908 3 C C
10 908 4 A A
| You are quite good my friend!
|========================================================= | 71%
| It's important to note that we left the id column in both tables. In the world of
| relational databases, 'id' is called our 'primary key' since it allows us to connect
| each student listed in student_info with their grades listed in gradebook. Without a
| unique identifier, we might not know how the tables are related. (In this case, we
| could have also used the name variable, since each student happens to have a unique
| name.)
...
|========================================================== | 73%
| The fifth and final messy data scenario that we'll address is when a single
| observational unit is stored in multiple tables. It's the opposite of the fourth
| problem.
...
|============================================================ | 75%
| To illustrate this, we've created two datasets, passed and failed. Take a look at
| passed now.
passed
name class final
1 Brian 1 B
2 Roger 2 A
3 Roger 5 A
4 Karen 4 A
| All that hard work is paying off!
|============================================================= | 76%
| Now view the contents of failed.
failed
name class final
1 Brian 5 C
2 Sally 1 C
3 Sally 3 C
4 Jeff 2 E
5 Jeff 4 C
6 Karen 3 C
| Your dedication is inspiring!
|=============================================================== | 78%
| Teachers decided to only take into consideration final exam grades in determining
| whether students passed or failed each class. As you may have inferred from the data,
| students passed a class if they received a final exam grade of A or B and failed
| otherwise.
...
|================================================================ | 80%
| The name of each dataset actually represents the value of a new variable that we will
| call 'status'. Before joining the two tables together, we'll add a new column to each
| containing this information so that it's not lost when we put everything together.
...
|================================================================= | 82%
| Use dplyr's mutate() to add a new column to the passed table. The column should be
| called status and the value, "passed" (a character string), should be the same for all
| students. 'Overwrite' the current version of passed with the new one.
passed<-mutate(passed,status="passed")
| That's a job well done!
|=================================================================== | 84%
| Now, do the same for the failed table, except the status column should have the value
| "failed" for all students.
failed<-mutate(failed,status="failed")
| Perseverance, that's the answer.
|==================================================================== | 85%
| Now, pass as arguments the passed and failed tables (in order) to the dplyr function
| bind_rows(), which will join them together into a single unit. Check ?bind_rows if you
| need help.
|
| Note: bind_rows() is only available in dplyr 0.4.0 or later. If you have an older
| version of dplyr, please quit the lesson, update dplyr, then restart the lesson where
| you left off. If you're not sure what version of dplyr you have, type
| packageVersion('dplyr').
bind_rows(passed,failed)
name class final status
1 Brian 1 B passed
2 Roger 2 A passed
3 Roger 5 A passed
4 Karen 4 A passed
5 Brian 5 C failed
6 Sally 1 C failed
7 Sally 3 C failed
8 Jeff 2 E failed
9 Jeff 4 C failed
10 Karen 3 C failed
| You nailed it! Good job!
|====================================================================== | 87%
| Of course, we could arrange the rows however we wish at this point, but the important
| thing is that each row is an observation, each column is a variable, and the table
| contains a single observational unit. Thus, the data are tidy.
...
|======================================================================= | 89%
| We've covered a lot in this lesson. Let's bring everything together and tidy a real
| dataset.
...
|========================================================================= | 91%
| The SAT is a popular college-readiness exam in the United States that consists of three
| sections: critical reading, mathematics, and writing. Students can earn up to 800
| points on each section. This dataset presents the total number of students, for each
| combination of exam section and sex, within each of six score ranges. It comes from the
| 'Total Group Report 2013', which can be found here:
|
| http://research.collegeboard.org/programs/sat/data/cb-seniors-2013
...
|========================================================================== | 93%
| I've created a variable called 'sat' in your workspace, which contains data on all
| college-bound seniors who took the SAT exam in 2013. Print the dataset now.
sat
# A tibble: 6 x 10
score_range read_male read_fem read_total math_male math_fem math_total write_male
1 700-800 40151 38898 79049 74461 46040 120501 31574
2 600-690 121950 126084 248034 162564 133954 296518 100963
3 500-590 227141 259553 486694 233141 257678 490819 202326
4 400-490 242554 296793 539347 204670 288696 493366 262623
5 300-390 113568 133473 247041 82468 131025 213493 146106
6 200-290 30728 29154 59882 18788 26562 45350 32500
# ... with 2 more variables: write_fem , write_total
| That's a job well done!
|============================================================================ | 95%
| As we've done before, we'll build up a series of chained commands, using functions from
| both tidyr and dplyr. Edit the R script, save it, then type submit() when you are
| ready. Type reset() to reset the script to its original state.
{r}
# Accomplish the following three goals:
#
# 1. select() all columns that do NOT contain the word "total",
# since if we have the male and female data, we can always
# recreate the total count in a separate column, if we want it.
# Hint: Use the contains() function, which you'll
# find detailed in 'Special functions' section of ?select.
#
# 2. gather() all columns EXCEPT score_range, using
# key = part_sex and value = count.
#
# 3. separate() part_sex into two separate variables (columns),
# called "part" and "sex", respectively. You may need to check
# the 'Examples' section of ?separate to remember how the 'into'
# argument should be phrased.
#
sat %>%
select(-contains("total")) %>%
gather(part_sex, count, -score_range) %>%
### <Your call to separate()> %>%
separate(part_sex,c("part","sex")) %>%
print
submit()
| Sourcing your script...
# A tibble: 36 x 4
score_range part sex count
1 700-800 read male 40151
2 600-690 read male 121950
3 500-590 read male 227141
4 400-490 read male 242554
5 300-390 read male 113568
6 200-290 read male 30728
7 700-800 read fem 38898
8 600-690 read fem 126084
9 500-590 read fem 259553
10 400-490 read fem 296793
# ... with 26 more rows
| You got it!
|============================================================================= | 96%
| Finish off the job by following the directions in the script. Save the script and type
| submit() when you are ready, or type reset() to reset the script to its original state.
{r}
# Append two more function calls to accomplish the following:
#
# 1. Use group_by() (from dplyr) to group the data by part and
# sex, in that order.
#
# 2. Use mutate to add two new columns, whose values will be
# automatically computed group-by-group:
#
# * total = sum(count)
# * prop = count / total
#
sat %>%
select(-contains("total")) %>%
gather(part_sex, count, -score_range) %>%
separate(part_sex, c("part", "sex")) %>%
### <Your call to group_by()> %>%
group_by(part,sex) %>%
mutate(total = sum(count),
prop = count / total
) %>% print
submit()
| Sourcing your script...
# A tibble: 36 x 6
# Groups: part, sex [6]
score_range part sex count total prop
1 700-800 read male 40151 776092 0.0517
2 600-690 read male 121950 776092 0.157
3 500-590 read male 227141 776092 0.293
4 400-490 read male 242554 776092 0.313
5 300-390 read male 113568 776092 0.146
6 200-290 read male 30728 776092 0.0396
7 700-800 read fem 38898 883955 0.0440
8 600-690 read fem 126084 883955 0.143
9 500-590 read fem 259553 883955 0.294
10 400-490 read fem 296793 883955 0.336
# ... with 26 more rows
| Keep up the great work!
|=============================================================================== | 98%
| In this lesson, you learned how to tidy data with tidyr and dplyr. These tools will
| help you spend less time and energy getting your data ready to analyze and more time
| actually analyzing it.
...
|================================================================================| 100%
| Would you like to receive credit for completing this course on Coursera.org?
1: Yes
2: No
Selection: 1
What is your email address? xxxxxx@xxxxxxxxxxxx
What is your assignment token? xXxXxxXXxXxxXXXx
Grade submission succeeded!
| Excellent job!
| You've reached the end of this lesson! Returning to the main menu...
| Please choose a course, or type 0 to exit swirl.
1: Getting and Cleaning Data
2: R Programming
3: Take me to the swirl course repository!
Selection: 0
| Leaving swirl now. Type swirl() to resume.
ls()
[1] "failed" "gradebook" "passed" "res" "sat"
[6] "student_info" "students" "students2" "students3" "students4"
rm(list=ls())